
Optimising your workflow is bound to be an ongoing task in an increasingly complex world, filled with increasingly complex data. In our blog, we discuss our quest for increased scalability and decreased runtimes in Python – including a little comparison of Dask vs. Spark, results from our Pandas/Dask experiment, and concluding with our final thoughts on what works best for our team here at Lynchpin.
Python is single-threaded. This is not an issue in most workloads, as a few extra seconds of runtime usually do not matter. However, in a world of ever-increasing datasets, some workloads can quickly increase in runtime – sometimes from a few minutes to several hours, or even days. In some instances, this results in a situation where data is generated faster than it is processed.
At Lynchpin, we have some workloads with a computation time of O(n2) which, when working with tens of millions of rows, gets unmanageable – QUICKLY. And not only does the run-time increase, running out of ram becomes a genuine concern. Working with bigger-than-ram datasets requires a careful approach and techniques like chunking where the dataset is split into several chunks which are processed individually. Doing this requires more development time as pre-processing scripts and chunking requires the developers to be more careful in their approach. This prompted us to move away from the tried and tested single-threaded python scripts and experiment with other solutions.
One solution to the issue of runtime would be to start working with the data in chunks and use the built-in multiprocessing library in Python to work on several chunks at the same time. This does however only do something to mitigate the increased runtimes and would only increase the risk of running out of ram. Furthermore, multiprocessing workloads often come with a few headaches as the script now must handle what each processor should work on and when. Making errors in this can result in a number of problems, including each processor thread spawning new processor threads until the computer becomes unresponsive.
This quest for scalability led us down several paths including parallelism, the topic of this blog.
When working with parallel computations, the program works on multiple calculations in parallel. This can be distinct tasks or working on chunks of the same task. While this can be done by multiple threads on a single core, that core can still only work on one task at a time and task switching comes with a time penalty which can result in increased runtime over single threaded processes (threading on a single core spawns a discussion on I/O and CPU bound tasks which is a little outside the scope of this blog).
Having the workload spread out between multiple CPU cores or even multiple CPUs is a much better approach as each core can focus on one task instead of switching between several concurrent tasks. With even commodity laptops getting more and more cores, being able to spread the workload across multiple cores becomes increasingly attractive. Once a computer is fully utilised, it is XXX to include other workers in the computation. When doing this, the task moves from being parallelised to being distributed. It is however very similar to parallelisation but is instead spread over several “workers”, each with its own processor and ram responsible for doing the computations and a scheduler that schedules and manages jobs submitted to the cluster and makes sure each worker node does what it is supposed to. Users can submit jobs to the scheduler which then makes sure they are run and returns the result. Most libraries and frameworks support both parallelisation and distribution.
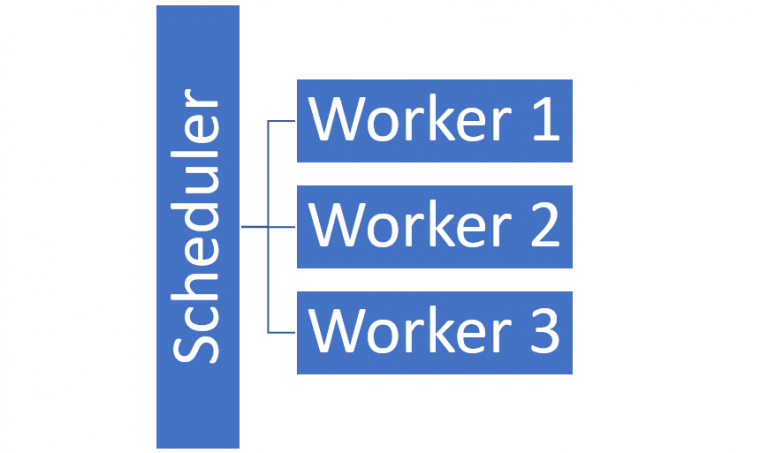
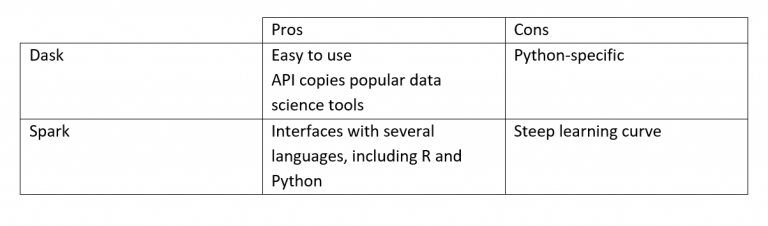
We decided to focus on Dask and Spark as Dask is a well-developed library and Spark is a widely used and well-documented framework that is interfaceable from languages widely used in the company. This blog will however mainly focus on Dask, but the results and considerations apply to other similar library and frameworks like Spark. Both Dask and Spark can scale from a local cluster (i.e. a single computer) to big multi-node clusters.
Dask is a python library adding parallelisation and the ability to distribute tools like Pandas, NumPy and SciKit-Learn. Most of the Dask API is a copy of the tools it tries to emulate which makes it easy to pick up and integrate into an existing workflow. A Dask DataFrame is just a bunch of smaller Pandas DataFrames distributed to the workers on the cluster and coordinated by Dask. Dask mirrors the functionality and API of Pandas which makes the switch easier than switching to e.g. Spark where most of the code would have to be rewritten.
Both solutions come with both pros and cons:
To test the improvement on runtime by switching to parallel computing, an experiment using the NYC Taxi Dataset will be set up. Using this, the great circle distance between the pickup and drop-off point will be calculated using the haversine formula and then calculate the average price per straight-line mile travelled across the dataset. The entire execution, from loading the dataset to calculating the average will be calculated. This could resemble a light analysis in a real-world scenario while still being relatively simple.
The experiment will be split up into two parts. The first part will be using Pandas’ .apply function (and the Dask equivalent) which applies a function across an axis in a DataFrame – in this case the columns. Using this, the haversine formula will be applied to each row of the DataFrame. Then the average price per mile travelled will be calculated by finding the price per trip and averaging these. This is the most common method used to apply functions to DataFrame but not the most effective. In the second part, an optimised version of the script will be used passing Pandas Series to the distance and price functions instead of individual numbers. Because most modern CPUs support vectorised calculations, this should result in a significant decrease in computation time, both for Pandas and Dask.
The two experiments will be run on two subsets of the Taxi dataset, 1 and 50 million rows, to emulate different workloads sizes. Everything will be done on an 8 core Xeon E5 and 64 GB ram.
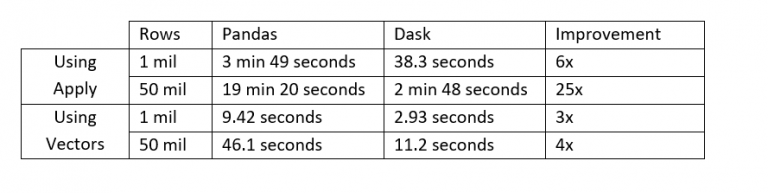
As seen in our results, moving to Dask resulted in a lower runtime in all scenarios. The greatest improvements were in the cases where apply was used, but in all cases, the runtime was decreased at least 3x by moving to Dask. The lower runtime improvements when using vectors is due to the time spent loading the dataset into memory.
Parallel or distributed computing is, however, not for everyone. Suddenly having more than one parallel process running results in increased overhead both from transferring data and files between cores on a local cluster or workers on the network and from collecting results from the cluster. As seen from the experiment, the biggest performance relative increase did not come from splitting the data into chunks and running it in parallel but simply shifting from using apply to doing vectorised computations on Pandas series. An even bigger increase could probably come from moving to vectorised computations over NumPy arrays, but this is not supported by Dask. Even though optimisation of the computation gave the biggest relative performance increase, moving to Dask is not necessarily a bad thing. Once a script is fully optimised and utilising the core it is running on, where can you go from there? That’s right – nowhere. And that is where a parallelised system comes into full force.
The processes in the experiment were embarrassingly parallelisable and vectorisable and not very memory intensive tasks. Had this been in the real world, it would be easy to expect that the data would need to be joined up with something like a driver database to predict car utilization or spikes in demand. Had a couple of joins been introduced or had we needed to train a model, “only” having 64 GB ram would quickly become an issue and a memory error would have been inevitable. Memory errors can be avoided but at the cost of increased complexity, development time and increased runtime. Situations like this are where a parallelised solution starts to really make a difference.
Instead of relying on a single core to do the work and scaling up once more power is needed (at an increasing marginal cost), a parallelised system can be scaled out by adding more relatively inexpensive nodes to the existing cluster. Equally, as Dask (and other parallelised systems) can easily work with larger-than-ram datasets by only reading what is needed and using a hard drive as excess, running out of memory is not a big issue. Shifting to parallel or distributed computing opens new possibilities by making it possible to process increasing amounts of data.
To find out more about our data engineering capabilities at Lynchpin, visit our services page. And be sure to follow us on Twitter and LinkedIn for all of our latest updates.
About the author
Jens Breuning
With a master’s degree specialising in business analytics, business process development, big data, platform strategies, AI and ML, Jens currently works as a Data Scientist for Lynchpin.
Jens specialises in BI projects utilising digital data as well as offline sources. He has a skill set that spans data engineering to analytics and is comfortable architecting data pipelines to support BI projects.
Jens works with a range of front-end BI and data visualisation tools such as Power BI and Tableau; He is familiar with many languages - including Python and SQL; Jens also works with cloud platforms Microsoft Azure and GCP (Google Cloud Platform).


